Microbial hydrogen metabolism
Like formate, hydrogen represents an important energy source for a multitude of microorganisms on this planet. Hydrogen produced by many fermenting microorganisms is also a means of removing redox equivalents from the cell. Hydrogen metabolism has also been a key driver in the evolution of life on earth. As such, the study of hydrogenases, the enzymes that reversibly activate hydrogen, is important not only in understanding the energetics of microbial growth, but also with regard to how they have rightly garnered biotechnological interest due to their potential use in alternative energy storage. Of these enzymes, the class of [NiFe]-hydrogenases is the most abundant and most ancient. [NiFe]-hydrogenases catalyze both oxidation of hydrogen, to provide reducing equivalents, and reduction of protons to generate hydrogen.
Our main research focus is understanding how the complex NiFe(CN)2CO cofactor is synthesized, assembled and introduced into the apo-form of the catalytic subunit of [NiFe]-hydrogenases. The biosynthesis of the [NiFe]-cofactor is of considerable interest because it represents a lucid example of coordinated metallo-cofactor assembly. Six Hyp proteins are involved in adding the one carbonyl- and two cyano-moieties to the Fe ion of the [NiFe]-cofactor. The Hyp proteins are highly conserved and required in all organisms that synthesize [NiFe]-hydrogenases. Identifying their specific roles in assembly of the cofactor is the main focus of our scientific research.
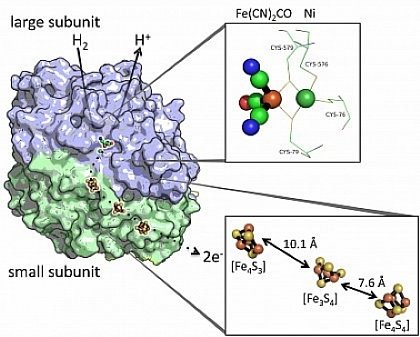
Hydrogenase 1: The large subunit in blue and small, iron-sulfur-containing subunit is represented in green.




