Links
Contact
Dr. Constanze Pinske
constanze.pinske@mikrobiolo...
room 217.1
Kurt-Mothes-Str. 3
06120 Halle (Saale)
postal address:
Dr. Constanze Pinske
06120 Halle (Saale)
Login for editors
Dr. Constanze Pinske
- Artikel über unsere Arbeit im 'Maulwurf' dem Blog des FSR Biologie Halle
- Uni Halle Magazin Campus Halensis Artikel über die Lange Nacht der Wissenschaften 2018
- MZ Halle Artikel Lokales 23.05.2017, page 12 by Sandy Schulze
- Scientia Halensis article about Constanze Pinske

Pinske (c) Kai Bublitz/L'Oréal
Curriculum Vitae - Constanze Pinske
| 2022 | Christian-Wolff Price for the Habilitation by the Martin-Luther-University Halle-Wittenberg |
| 2021 | Habilitation at the Martin-Luther-University Halle-Wittenberg |
| 2017 | “For Women in Science“ fellowship of the German UNESCO-Commission, L’Oréal Germany and the Christiane Nüsslein-Volhard-Stiftung |
| since 10/2015 | Division of General Microbiology, Martin-Luther University/Germany |
| 2012-2015 | School of Life Sciences, University of Dundee/UK; Postdoctoral research assistant in the group of Prof. Frank Sargent |
| 2008-2011 | PhD thesis ’Maturation and Assembly of [NiFe]-hydrogenases in Escherichia coli’ at the Division of General Microbiology Martin-Luther University Halle-Wittenberg, Germany under the supervision of Prof. R. Gary Sawers; viva passed with summa cum laude. |
| 2007-2008 | Diploma thesis ’Maturation of [NiFe]-hydrogenases strategies for isolating hydrogenase negative mutants in Escherichia coli’ at the Division of General Microbiology Martin-Luther University Halle-Wittenberg, Germany under the supervision of Prof. R. Gary Sawers |
| 2003-2008 | Martin-Luther University/Germany; Diploma in Biochemistry |
| 1983 | born in Berlin |
Research
Escherichia coli is a facultative anaerobic enterobacterium that can grow using oxygen as terminal electron acceptor during aerobic respiration. Alternatively, and in the absence of oxygen, it uses a variety of different electron acceptors or it ferments sugars (Ingledew and Poole, 1984; Sawers and Clark, 2004). Fermentable sugars are metabolized to pyruvate via glycolysis and subsequently converted to acetyl-CoA and formate due to the action of the key enzyme of mixed acid fermentation pyruvate formate lyase (PflB; Figure 1A). It is assumed that formate is toxic for the bacterial cell and thus is readily exported by e.g. the formate transporter FocA (Sawers, 2005). However, at the beginning of the stationary phase the cells re-import formate as substrate for the formate hydrogenlyase (FHL) complex, whereby it is oxidized to CO2 and H2 (Figure 1B).
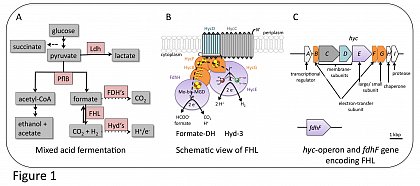
Hydrogen is an attractive renewable fuel and thus the FHL as the main H2-producing enzyme complex in E. coli is a potential candidate for biotechnological H2 production. The FHL complex is membrane-bound and comprises seven subunits (Sauter et al., 1992; McDowall et al., 2014). The formate dehydrogenase (FDH-H) is the molybdenum/selenium containing subunit that catalyses the oxidation of formate into CO2 and electrons. Those electrons are transferred to the hydrogenase catalytic subunit (HycE) via three iron-sulfur cluster containing subunits HycB, HycF and HycG. At HycE they are used to reduce protons to form H2. HycE harbours the typical hydrogenase [NiFe]-active site cofactor. The five subunits FDH-H-HycBEFG form the soluble domain of FHL, which in addition associates with two membrane subunits (HycC and HycD). Although there is no evidence that these membrane subunits participate in the catalytic reaction, they are nevertheless required for electrochemical coupling of the FDH-H and HycE reactions.
Due to its inherent structural similarities to complex I of the respiratory chain (Figure 2), it has been assumed that FHL also has an energy-conserving function (Böhm et al., 1990; Trchounian and Sawers, 2014; McDowall et al., 2014). However, this has not yet been experimentally proven (Pinske and Sargent, 2016). It is conceivable that energy conservation is only achieved by interaction with other membrane proteins.

Hydrogen car shows a possible application for hydrogen as fuel after conversion
into electricity by a fuel cell.
Although the HycE protein has previously served as a model for maturation, which includes the biosynthesis and insertion of its [NiFe]-cofactor, next to nothing is known about the subsequent steps in hydrogenase assembly. Some research in this direction has been done with the periplasmically-oriented hydrogenases of E. coli (Dubini and Sargent, 2003); however assembly of the cytoplasmically-oriented FHL complex is dependent on further proteins whose functions are not yet understood. Research in my group aims at understanding the assembly of FHL, the unique proteins involved in this process and the involvement of FHL in the cellular metabolism and bioenergetics. Understanding FHL will aid the further development of this complex for biotechnological applications.
Bibliography
Böhm, R., Sauter, M., and Böck, A. (1990) Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol Microbiol 4: 231–243.
Dubini, A., and Sargent, F. (2003) Assembly of Tat-dependent [NiFe] hydrogenases: identification of precursor-binding accessory proteins. FEBS Lett 549: 141–146.
Ingledew, W., and Poole, R. (1984) The respiratory chains of Escherichia coli. Microbiol Rev 48: 222–271.
McDowall, J.S., Murphy, B.J., Haumann, M., Palmer, T., Armstrong, F.A., and Sargent, F. (2014) Bacterial formate hydrogenlyase complex. Proc Natl Acad Sci USA 111: E3948–56.
Pinske, C. and Sargent, F. (2016) Exploring the directionality of Escherichia coli formate hydrogenlyase: a membrane-bound enzyme capable of fixing carbon dioxide to organic acid. MicrobiologyOpen 5, 721–737.
Sauter, M., Böhm, R., and Böck, A. (1992) Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol Microbiol 6: 1523–1532.
Sawers, R.G. (2005) Formate and its role in hydrogen production in Escherichia coli. Biochem Soc Trans 33: 42–46.
Sawers, R.G., and Clark, D. (2004) Fermentative Pyruvate and Acetyl-Coenzyme A Metabolism. EcoSal - Escherichia coli and Salmonella: cellular and molecular biology.
Trchounian, A., and Sawers, R.G. (2014) Novel insights into the bioenergetics of mixed-acid fermentation: Can hydrogen and proton cycles combine to help maintain a proton motive force? IUBMB Life 66: 1–7.
Publications
1. Kammel M, Trebbin O, Pinske C and Sawers RG*. 2021. A single amino acid exchange converts FocA into a unidirectional efflux channel for formate. Microbiology (in press) doi: 10.1099/mic.0.001132
2. Telleria Marloth J and Pinske C*. 2020. Susceptibility of the formate hydrogenlyase reaction to the protonophore CCCP depends on the total hydrogenase composition. Inorganics, 8, 38.
3. Karapetyan L, Pinske C, Sawers RG, Trchounian A, Trchounian K*. 2020. Influence of C4 -Dcu transporters on hydrogenase and formate dehydrogenase activities in stationary phase-grown fermenting Escherichia coli. IUBMB Life 72: 1680–1685.
4. Skorupa P, Lindenstrauß U, Burschel S, Blumenscheit C, Friedrich T, Pinske C*. 2020. The N-terminal domains of the paralogous HycE and NuoCD govern assembly of the respective formate hydrogenlyase and NADH dehydrogenase complexes. FEBS Open Bio 10:371–385.
5. Pinske C, Thomas C, Nutschan K and Sawers RG*. 2019. Delimiting the function of the C-terminal extension of the Escherichia coli [NiFe]-hydrogenase 2 large subunit precursor. Front. Microbiol. 10:2223.
6. Lubek D, Simon AH, Pinske C*. 2019. Amino acid variants of the HybB membrane subunit of Escherichia coli NiFe-hydrogenase-2 support a role in proton translocation. FEBS Letters 156:277.
7. Lindenstrauß U, Pinske C*. 2019. Dissection of the hydrogen metabolism of the enterobacterium Trabulsiella guamensis: Identification of a formate-dependent and essential formate hydrogenlyase complex exhibiting phylogenetic similarity to complex I. J. Bacteriol. 201(12):1480.
Preprint: BioRxiv https://doi.org/10.1101/563510
8. Pinske C*. 2019. Bioenergetic aspects of archaeal and microbial hydrogen metabolism. Adv Microb Physiol vol. 74:487-514.
9. Dragomirova N, Rothe P, Schwoch S, Hartwig S, Pinske C*, Sawers RG*. 2018. Insights into the redox sensitivity of Chloroflexi Hup-hydrogenase derived from studies in Escherichia coli: Merits and pitfalls of heterologous NiFe-hydrogenase synthesis. Front Microbiol. 9:2837.
10. Hakobyan B, Pinske C, Sawers RG, Trchounian A, Trchounian K*. 2018. PH and a mixed carbon-substrate spectrum influence FocA- and FocB-dependent, formate-driven H2 production in Escherichia coli. FEMS Microbiol Lett. 365(21)
11. Pinske C*. 2018. The ferredoxin-like proteins HydN and YsaA enhance redox dye-linked activity of the formate dehydrogenase H component of the formate hydrogenlyase complex. Frontiers Microbiol. 9:1238
12. Thomas C, Waclawek M, Nutschan K, Pinske C, Sawers RG*. 2018 The extended C-terminal alpha-helix of the HypC chaperone restricts recognition of large subunit precursors by the Hyp-scaffold machinery during [NiFe]-hydrogenase maturation in Escherichia coli. J. Mol. Microbiol. Biotechnol. 28(2):87-97.
13. Lindenstrauß U, Skorupa P, McDowall JS, Sargent F, Pinske C*. 2017. The dual-function chaperone HycH improves assembly of the formate hydrogenlyase complex. Biochem J. 474:2937-2950.
14. Jaroschinsky M, Pinske C, Sawers RG*. 2017. Differential effects of isc operon mutations on the biosynthesis and activity of key anaerobic metalloenzymes in Escherichia coli. Microbiology (Reading, Engl.). 163:878-890.
15. Lamont CM, Kelly CL, Pinske C, Buchanan G, Palmer T, Sargent F*. 2017. Expanding the substrates for a bacterial hydrogenlyase reaction. Microbiology (Reading, Engl). 163:649-653.
16. (Book chapter) Sawers RG*, Pinske C (2017) “Insights into [NiFe]-Hydrogenase Active Site Metallocluster Assembly”, in ‘Metalloprotein Active Site Assembly’, edited by Michael K. Johnson and Robert A. Scott. Chichester, UK: John Wiley & Sons, Ltd, pp. 1-11.
17. (Review) Pinske C*, Sawers RG*. 2016. Anaerobic formate and hydrogen metabolism. EcoSal Plus. doi: 10.1128/ecosalplus.ESP-0011-2016
18. Pinske C, Sargent F*. 2016. Exploring the directionality of Escherichia coli formate hydrogenlyase: a membrane-bound enzyme capable of fixing carbon dioxide to organic acid. MicrobiologyOpen. 5:721-737
19. Kelly CL, Pinske C, Murphy BJ, Parkin A, Armstrong F, Palmer T, Sargent F*. 2015. Integration of an [FeFe]-hydrogenase into the anaerobic metabolism of Escherichia coli. Biotechnology Reports. 8:94-104
20. Hartwig S, Pinske C, Sawers RG*. 2015. Chromogenic assessment of the three molybdo-selenoprotein formate dehydrogenases in Escherichia coli. Biochemistry and Biophysics Reports. 1:62–67
21. Pinske C*, Sargent F, Sawers RG. 2015. SlyD-dependent nickel delivery limits maturation of [NiFe]-hydrogenases in late-stationary phase Escherichia coli cells. Metallomics. 7:683–90
22. Pinske C, Jaroschinsky M, Linek S, Kelly CL, Sargent F, Sawers RG*. 2015. Physiology and bioenergetics of [NiFe]-hydrogenase 2-catalyzed H2-consuming and H2-producing reactions in Escherichia coli. J Bacteriol. 197:296–306
23. (Review) Pinske C, Sawers RG*. 2014. The importance of iron in the biosynthesis and assembly of [NiFe]-hydrogenases. Biomol Concepts. 5:55–70
24. Pinske C, Jaroschinsky M, Sawers RG*. 2013. Levels of control exerted by the Isc iron-sulfur cluster system on biosynthesis of the formate hydrogenlyase complex. Microbiology (Reading, Engl). 159:1179–89
25. Trchounian K, Pinske C, Sawers RG, Trchounian A*. 2012. Characterization of Escherichia coli [NiFe]-hydrogenase distribution during fermentative growth at different pHs. Cell Biochem Biophys. 62:433–40
26. Pinske C, McDowall JS, Sargent F, Sawers RG*. 2012. Analysis of hydrogenase 1 levels reveals an intimate link between carbon and hydrogen metabolism in Escherichia coli K-12. Microbiology (Reading, Engl). 158:856-868
27. Pinske C, Sawers RG*. 2012. A-type carrier protein ErpA is essential for formation of an active formate-nitrate respiratory pathway in Escherichia coli K-12. J Bacteriol. 194:346–53
28. Pinske C, Sawers RG*. 2012. Delivery of iron-sulfur clusters to the hydrogen-oxidizing [NiFe]-hydrogenases in Escherichia coli requires the A-type carrier proteins ErpA and IscA. PLoS ONE. 7:e31755
29. Pinske C, Jaroschinsky M, Sargent F, Sawers RG*. 2012. Zymographic differentiation of [NiFe]-hydrogenases 1, 2 and 3 of Escherichia coli K-12. BMC Microbiol. 12:134
30. Pinske C, Krüger S, Soboh B, Ihling C, Kuhns M, Braussemann M, Jaroschinsky M, Sauer C, Sargent F, Sinz A, Sawers RG*. 2011. Efficient electron transfer from hydrogen to benzyl viologen by the [NiFe]-hydrogenases of Escherichia coli is dependent on the coexpression of the iron-sulfur cluster-containing small subunit. Arch Microbiol. 193:893–903
31. Trchounian K, Pinske C, Sawers RG, Trchounian A*. 2011. Dependence on the F0F1-ATP synthase for the activities of the hydrogen-oxidizing hydrogenases 1 and 2 during glucose and glycerol fermentation at high and low pH in Escherichia coli. J. Bioenerg. Biomembr. 43:645–50
32. Pinske C, Sawers RG*. 2011. Iron restriction induces preferential down-regulation of H2-consuming over H2-evolving reactions during fermentative growth of Escherichia coli. BMC Microbiol. 11:196
33. Pinske C, Bönn M, Krüger S, Lindenstrauß U, Sawers RG*. 2011. Metabolic deficiences revealed in the biotechnologically important model bacterium Escherichia coli BL21(DE3). PLoS ONE. 6:e22830
34. Soboh B, Pinske C, Kuhns M, Waclawek M, Ihling C, Trchounian K, Trchounian A, Sinz A, Sawers RG*. 2011. The respiratory molybdo-selenoprotein formate dehydrogenases of Escherichia coli have hydrogen: benzyl viologen oxidoreductase activity. BMC Microbiol. 11:173
35. Soboh B, Krüger S, Kuhns M, Pinske C, Lehmann A, Sawers RG*. 2010. Development of a cell-free system reveals an oxygen-labile step in the maturation of [NiFe]-hydrogenase 2 of Escherichia coli. FEBS Lett. 584:4109–14
36. Pinske C, Sawers RG*. 2010. The role of the ferric-uptake regulator Fur and iron homeostasis in controlling levels of the [NiFe]-hydrogenases in Escherichia coli. International Journal of Hydrogen Energy. 35:8938–44
* corresponding author

